Cellular pathways represent the intricate metabolic connections between the plethora of signaling pathways, and energetic-sensors in organs. To maintain energy balance in body organs these metabolic pathways must act uniformly at any given point in time. This is to ensure organ functionality under specific metabolic changes. A new class of non-coding RNAs, MicroRNA, has recently been discovered which has proven the process of metabolic homeostasis to be more complex than it was previously thought.
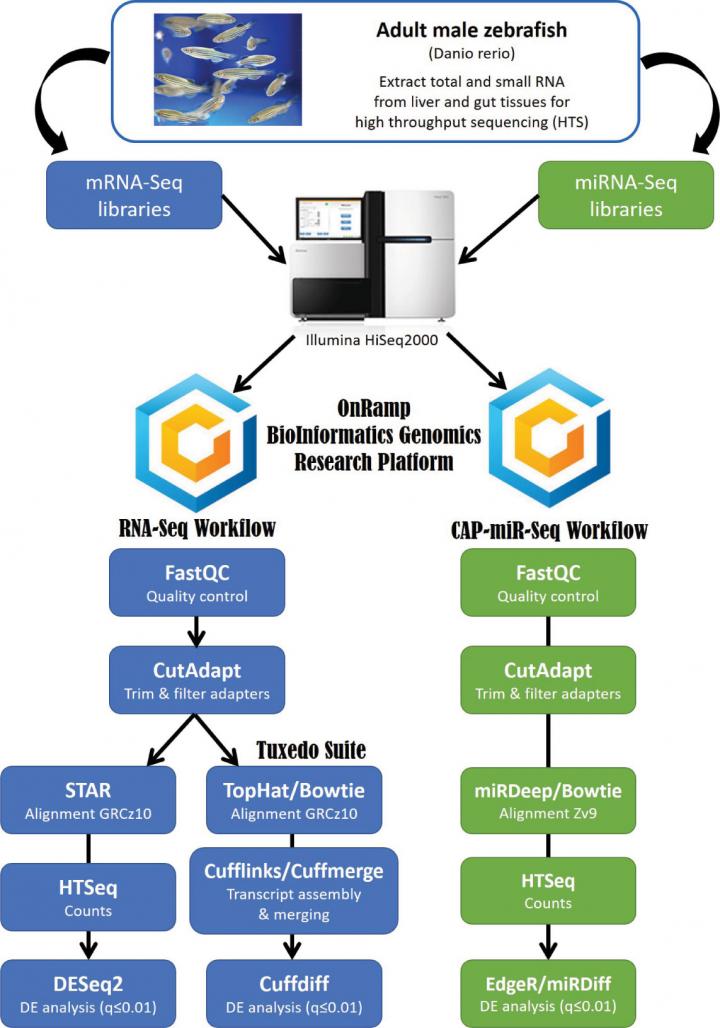
The objective of the research is to understand damages resulted by toxins to the liver and the intestine as well as its relation to MiRNome. Baseline Characterization in healthy animal tissue undergoing cellular homeostasis is required for initiating transcriptome process. For the experiment, researchers dissected wildtype fish (Zebrafish) and isolated their liver and gut; from these organs, small RNA was extracted. These organs were chosen because the liver is the main site for detoxification and gut is the primary site for contaminant exposure. From the RNA samples, mRNA and miRNA libraries were constructed and put under high throughout sequencing. Following sequencing, differential analysis was performed comparing liver mRNA contents with those in the gut. An “miRNA matrix” containing miRNA sequence and mRNA sequence was then constructed.
The results from the analysis have provided new information regarding microRNA function in these tissues. The miRNome and transcriptome of both liver and gut tissues were characterized and the related miRNAs were identified. Studying the miRNA matrix revealed two types of miRNA in liver and gut. miRNAs unique to each organ regulate fundamental cellular process important for both organs while those common to both tissues regulate biological processed specific to either the liver or the gut.

