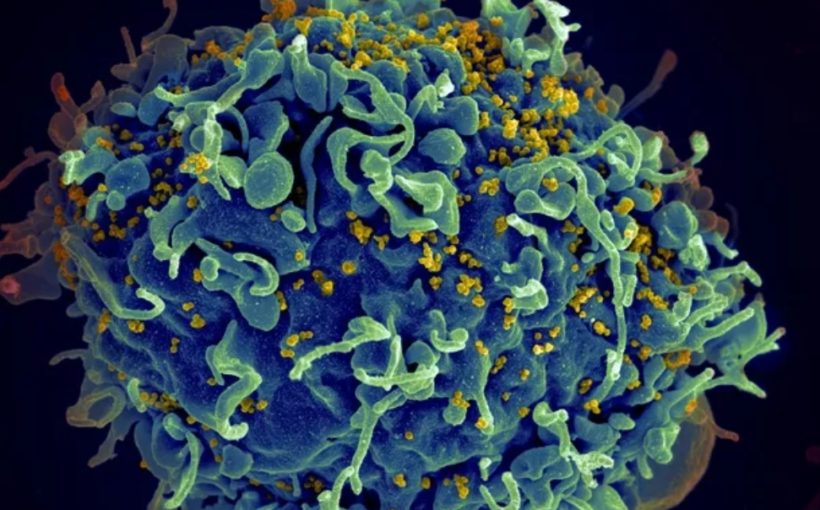
This newly-identified mechanism could open up a new avenue for control of HIV, the virus that causes AIDS. Most of the genome is made up of noncoding RNAs which do not directly translate into proteins. In fact, 97% of the human genome is non-protein coding. Dr. Kulkarni says that until the last decade or so, scientists thought many of these particular RNAs were “junk.” Now, new research shows they play many roles.
Recent developments in technology and genomics have made advances in knowledge in this area possible. Dr. Kulkarni and her colleagues showed that a specific long noncoding RNA impacts the gene encoding the HIV co-receptor CCR5. Since CCR5 is critical for the HIV virus to enter the cell, a polymorphism associated with variation in expression of this long noncoding RNA impact the infection’s outcome.
Genomic DNA from various groups including Hispanics, African Americans and Japanese showed that this association is present across many ethnicities. This consistency of this association across populations speak to a single functional mechanism explaining this association.
Nature Immunology also featured a commentary about the study called “A SNP of lncRNA gives HIV-1 a boost” to further underscore the importance of this study to the field. In the article, Sanath Kumar Janaka and David T. Evans write “this study provides new insight into how polymorphisms in regulatory elements may explain genetic variation in pathogenesis.” They go on to call Dr. Kulkarni’s discovery a “fascinating and intricate mechanism.”
“Their comments are encouraging and impel us to explore further,” Dr. Kulkarni said. “HIV is still a major public health burden. More than a million people are living with HIV in the U.S. alone. More than 50,000 new cases are reported each year. “Finding functional mechanisms of the disease-associated gene regions will increase our understanding about how they regulate disease-associated genes and pathways,” Dr. Kulkarni explained. “We may be able to find selective targets for therapy.”