The antibody discovery and vaccine development research fields may be on the verge of rapidly expanding with data that previously took decades to acquire, thanks to LIBRA-seq, a new tool developed by Vanderbilt University researchers and their colleagues.
Currently, due to technological constraints, it can take up to a year to identify—from the billions of antibodies produced by white blood cells called B cells—a single antibody that can be developed into a drug that recognizes and attacks the antigen marker of a specific pathogen.
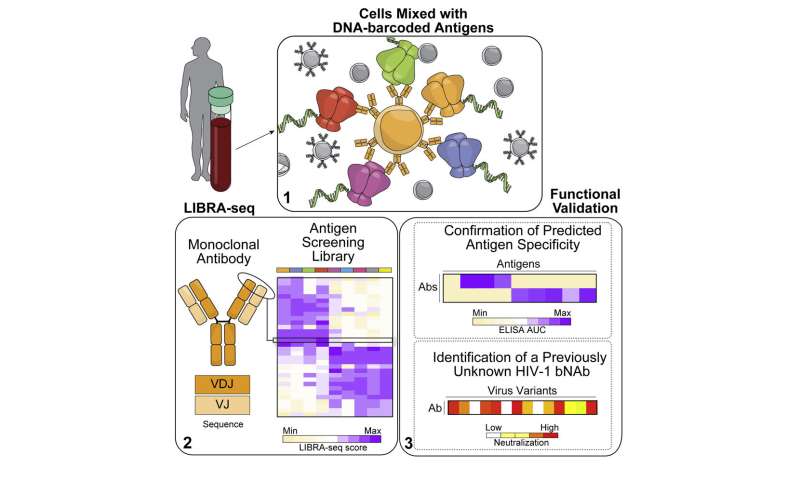
Ian Setliff, a graduate student in the lab of Ivelin Georgiev, Ph.D., in the Vanderbilt Vaccine Center, wondered if it was possible to map antibody sequences and antigen specificities simultaneously and in a high throughput way.
“That seemed like a crazy idea,” said Georgiev, “something that very likely won’t work.” But Setliff’s concept did work and led to the discovery of a novel antibody against HIV in just weeks.
The development and validation of LIBRA-seq, which stands for linking B-cell Receptor to antigen specificity through sequencing, was described recently in the journal Cell.
Normally “it’s very hard to know when you only have the antibody sequence, what the antigen specificity of that antibody is,” said Georgiev, associate professor of Pathology, Microbiology and Immunology at Vanderbilt University Medical Center and associate professor of Computer Science in the Vanderbilt University School of Engineering.
Working with colleagues from Duke University School of Medicine, Massachusetts General Hospital, the University of Witwatersrand in Johannesburg, South Africa, and the National Institutes of Health, which supported the effort, the Vanderbilt researchers began by validating LIBRA-seq on cell lines with known B cell receptors.
Then they moved to prospective antibody discovery, which identified a previously unknown lineage of broadly neutralizing antibodies against the human immunodeficiency virus (HIV), which causes AIDS.
“The method really facilitates discovery because we were able to use a much larger screening library than would ever be possible with other screening methods,” said Vanderbilt graduate student Andrea Shiakolas, who shared the paper’s first-author status with Setliff.
The multi-institution collaboration and resources available at Vanderbilt made the complex project possible, Georgiev said.
Those resources included Vanderbilt Technologies for Advanced Genomics (VANTAGE), VUMC’s core genomics laboratory, the medical center’s Flow Cytometry Shared Resource and the university’s Advanced Computing Center for Research and Education (ACCRE).
“We’ve been very fortunate to be able to benefit from fantastic collaborations both here at Vanderbilt as well as outside of Vanderbilt,” Georgiev said, highlighting the resources of the VANTAGE sequencing core as essential and critical for the success of the project.
The team is hopeful that LIBRA-seq will be a transformative technology that will have a direct impact on both basic and applied research. “It enables us to start asking a lot more questions that we were previously unable to ask,” he said.
“We’re starting to study new targets within the HIV infection and vaccination field, but we’re also interested in the basic biology of how antibodies recognize antigens,” Georgiev added. “This technology will help us generate a lot more data that we’ll be able to mine to start addressing these questions.”
LIBRA-seq has the potential to help researchers narrow their search to find targets that could profoundly impact human health, the researchers said.
It’s a feat comparable to trying to find a few special stars out of the billions that make up the universe, they said. LIBRA-seq’s name, after all, was inspired by Libra, the astronomical constellation.
Vanderbilt University

