The recent coronavirus (COVID-19) outbreak presents enormous challenges for global health. To aid the global effort, Broad Institute of MIT and Harvard, the McGovern Institute for Brain Research at MIT, and our partner institutions have committed to freely providing information that may be helpful, including by sharing information that may be able to support the development of potential diagnostics.
The recent coronavirus (COVID-19) outbreak presents enormous challenges for global health. To aid the global effort, Broad Institute of MIT and Harvard, the McGovern Institute for Brain Research at MIT, and our partner institutions have committed to freely providing information that may be helpful, including by sharing information that may be able to support the development of potential diagnostics.
As part of this effort, Feng Zhang, Omar Abudayyeh, and Jonathan Gootenberg have developed a research protocol, applicable to purified RNA, that may inform the development of CRISPR-based diagnostics for COVID-19.
This initial research protocol is not a diagnostic test and has not been tested on patient samples. Any diagnostic would need to be developed and validated for clinical use and would need to follow all local regulations and best practices.
The research protocol provides the basic framework for establishing a SHERLOCK-based COVID-19 test using paper strips.
The team welcomes researchers to contact them for assistance or guidance and can provide a starter kit to test this system, as available, for researchers working with COVID-19 samples.
The SHERLOCK protocol
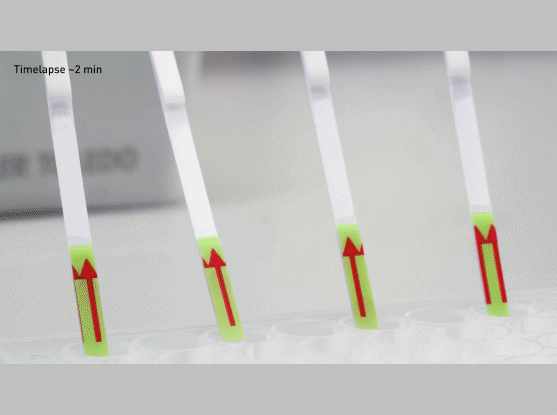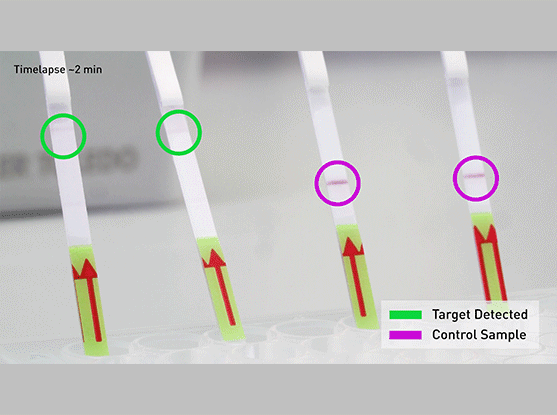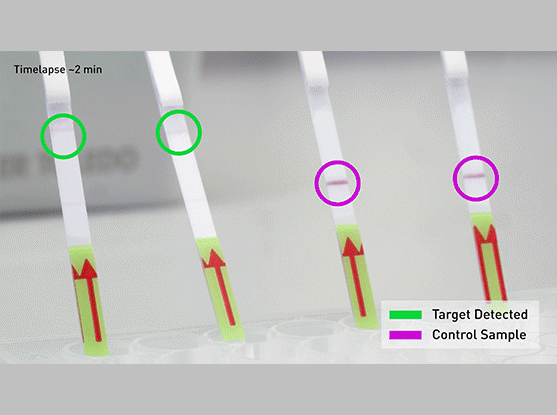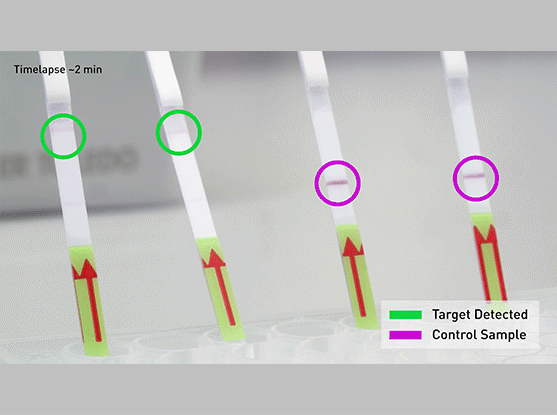
The CRISPR-Cas13-based SHERLOCK system has been previously shown to accurately detect the presence of a number of different viruses in patient samples. The system searches for unique nucleic acid signatures and uses a test strip similar to a pregnancy test to provide a visual readout. After dipping a paper strip into a prepared sample, a line appears on the paper to indicate whether the virus is present.
Using synthetic COVID-19 RNA fragments, the team designed and tested two RNA guides that recognize two signatures of COVID-19. When combined with the Cas13 protein, these form a SHERLOCK system capable of detecting the presence of COVID-19 viral RNA.
The research protocol involves three steps. It can be used with the same RNA samples that have been extracted for current qPCR tests:
- Incubate extracted RNA with isothermal amplification reaction for 25 min at 42C
- Incubate reaction from step 1with Cas13 protein, guide RNA, and reporter molecule for 30 min at 37C
- Dip the test strip into reaction from step 2, and result should appear within five minutes.
Further details which researchers and laboratories can follow (including guide RNA sequences), can be found in the .pdf protocol, which is available here and has been submitted to bioRxiv. The protocol will be updated as the team continues experiments in parallel and in partnership with those around the world seeking to address this outbreak. The researchers will continue to update this page with the most advanced solutions.
Necessary plasmids are available through the Zhang Lab Addgene repository, and other materials are commercially available. Details for how to obtain these materials are described in the protocol.
What’s next
The SHERLOCK diagnostic system has demonstrated success in other settings. The research team hopes the protocol is a useful step towards creating a system for detecting COVID-19 in patient samples using a simple readout. Further optimization, production, testing, and verification are still needed. Any diagnostic would need to follow all local regulations, best practices, and validation before it could become of actual clinical use. The researchers will continue to release and share protocol updates, and welcome updates from the community.
Broad Institute of MIT and Harvard