Clinicians have historically used family health history collection as a primary method for identifying actionable disease-risk assessments. However, as large-scale genome screening programs continue to rise in popularity, the scientific community has questioned the efficacy of this traditional method, wondering at the superiority of variants identified by sequencing over family health history in identifying these risks.
In order to address this question, the Duke Center for Applied Genomics and Precision Medicine (CAGPM) recently collaborated with the SingHealth Duke-National University of Singapore (NUS) Institute of Precision Medicine (PRISM) to evaluate how family health history can best synergize with genomic sequencing. Their findings were recently published in Genome Medicine.
In this study, CAGPM faculty members Geoffrey Ginsburg, MD, Ph.D., professor of medicine, (Cardiology), Lori Orlando, MD, associate professor of medicine (General Internal Medicine), Ryanne Wu, MD, assistant professor of medicine, (General Internal Medicine) and the PRISM team, discovered that integrating pre-emptive family health history collection in addition to genomic screening resulted in a higher likelihood of detecting carriers for cancer syndromes.
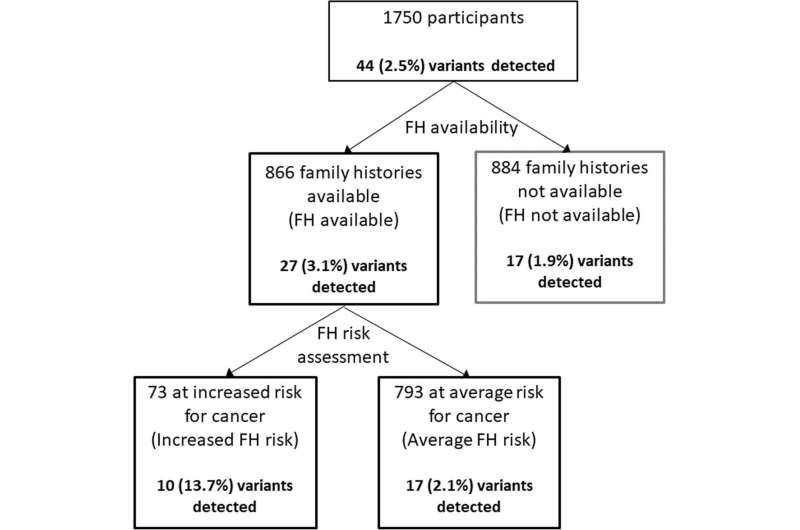
The study, conducted in Singapore, included 1750 participants with no known pre-existing medical conditions who consented to both a medical and genetic screen through PRISM.
“This study not only shows the impact of family history on diverse populations in South East Asia but the power of its use to prioritize genome sequencing in optimizing risk assessment” said Dr. Ginsburg.
Where PRISM lacked a mechanism for comprehensive family health history collection, CAGPM’s MeTree, an online tool concepted for this purpose, provided an avenue thorough enough for scientific data collection, but facile enough for patient use.
The study found that among 866 people whose family histories were collected, 73 were considered at risk of developing cancer and one in seven had a dominant clinically actionable variant. This is a six-fold increase compared to one in 47 of the 793 considered not to have an increased risk of developing cancer, and a sevenfold increase to one in 52 of those with no family health history. When assessing only the 25 cancer genes in the American College of Medical Genetics (ACMG) Secondary Findings list, those with a significant family health history were 18 times as likely to have a disease-causing variant compared to those without significant family health history.
63 participants with a significant family health history were not found to have a disease-causing variant.
Dr. Orlando cited several reasons for why a genomic health screen may miss risks identified by family health, “Family health history represents the combination of shared genes and shared environment. Thus, when the shared environment is the sole or primary contributor to a disease risk, you cannot see it with just a genomic test,” said Orlando. “Genomics is a rapidly evolving field and we still have much to learn. A family could plausibly have a pathogenic variant that has not yet been identified by science.”
For clinicians, family health history implementation is useful in guiding patient expectations when discussing the probability of detecting disease-causing variants. After detection, family history data, coupled with genotype, can provide patients with more precise indications of disease penetrance.
“What this study shows us is that family health history remains an important first-line screening tool in identifying people most likely to be at risk for a hereditary cancer syndrome,” said Dr. Wu.
This area of research, along with the collaboration between Duke-NUS and CAGPM, is just beginning. Investigators intend to expand to larger, more diverse populations across the globe, and are interested in focusing on under-served communities who are often left with insufficient resources to proactively address their health risks.
Milla Surjadi, Duke University School of Nursing

