Making memories involves more than seeing friends or taking photos. The brain constantly adapts to new information and stores memories by building connections among neurons, called synapses. How neurons do this—reaching out arm-like dendrites to communicate with other neurons—requires a ballet of genes, signaling molecules, cellular scaffolding and protein-building machinery.
A new study from scientists at Scripps Research and the Max Planck Florida Institute for Neuroscience finds a central role for one signaling molecule, a long, noncoding RNA that the scientists named ADEPTR.
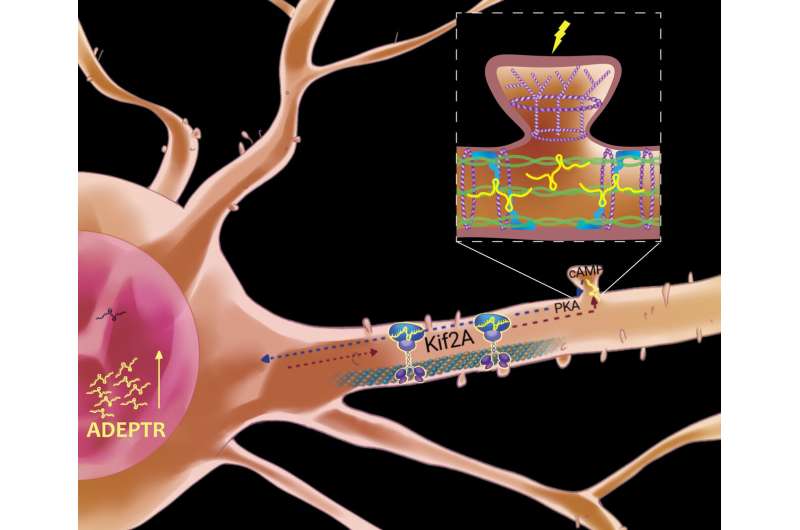
Using a variety of technologies, including confocal and two-photon microscopy, they track ADEPTR’s moves, watching as it forms, travels, amasses at the synapse and activates other proteins upon a neuron’s stimulation.
Its journey to the far reaches of a brain cell is made possible by a cellular carrier that that tiptoes along a dendrite’s microtubule scaffolding. Called a kinesin motor, it deposits ADEPTR near the synapse junction, where it activates other proteins.
The team also found that if ADEPTR is silenced, new synapses don’t form during stimulation.
The study, “Activity regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites,” is published online April 16 in the journal Science Advances.
Long noncoding RNAs have often been described as “genomic dark matter,” because their role in cells has yet to be fully characterized, especially in neurons, says the study’s lead author, Scripps Research neuroscientist Sathyanarayanan Puthanveettil, Ph.D. Puthanveettil’s team is finding that they play a signaling role in neural plasticity—how neurons adapt and change with experience.
“Here we report activity-dependent dendritic targeting of a newly transcribed long noncoding RNA for modulating synapse function, and describe its underlying mechanisms,” Puthanveettil says. “These studies bring novel insights into the functions of long noncoding RNAs at the synapse.”
The first author is Eddie Grinman, a graduate student in Puthanveettil’s lab.
A long noncoding RNA is a type of RNA that exceeds 200 nucleotides, and does not get translated into protein. There are thousands of these long noncoding RNA in our cells, but in most cases, their function isn’t yet known. What is known is that usually, they tend to stay within the cell nucleus. Some regulate the transcription of genes.
“It was surprising to see a long noncoding RNA move from nucleus to the synapse so rapidly and robustly,” Grinman says.
The hippocampus is the part of the brain where learning, memory and emotions reside. Working in hippocampal neurons from mice, the team stimulated the neurons with pharmacological activators of learning-related signaling. They found through molecular and high-resolution imaging techniques that the ADEPTR long noncoding RNA was rapidly expressed and transported to the outer arms of the cell. There, the ADEPTR molecules interact with proteins that play a role in structural organization of synapses, proteins called spectrin 1 and ankyrin B.
They found that ADEPTR became downregulated if exposed to an inhibitory neurotransmitter, GABA.
“These findings add another layer of complexity in synapse modulation and plasticity,” Puthanveettil says. “Synaptically localized long noncoding RNA are important regulators of adaptive neuronal function.”
Going forward, the team intends to continue characterizing how stimulation affects neuronal plasticity. Also, the authors hope to learn more about the role of ADEPTR in vivo.
“It would be interesting to learn what role ADEPTR plays in forming new memories in living organisms,” says Grinman.
The work is revealing one of the most fundamental processes of learning and memory, adaptation to changing information and circumstances.
“Neural plasticity is what allows us to learn, respond to stimuli, and lay down long-term memories,” Puthanveettil says. “There is still much to learn about the magnificent complexity of this fundamental biological process.”
The Scripps Research Institute

