Christopher Plaisier, an assistant professor of biomedical engineering in the Ira A. Fulton Schools of Engineering at Arizona State University, and Samantha O’Connor, a biomedical engineering doctoral student in the Plaisier Lab, are leading research into a new stage of the stem cell life cycle that could be the key to unlocking new methods of brain cancer treatment. Their work was recently published in the research journal Molecular Systems Biology.
“The cell cycle is such a well-studied thing and yet here we are looking at it again for the umpteenth time and a new phase pops out at us,” Plaisier says. “Biology always has new insights to show us, you just have to look.”
The spark for this discovery came through a collaboration with Patrick Paddison, an associate professor at the Fred Hutchinson Cancer Research Center in Seattle, and Dr. Anoop Patel, an assistant professor of neurological surgery at the University of Washington who is also involved in the Fred Hutchinson Cancer Research Center.
Paddison’s team called upon Plaisier to help analyze their brain stem cell data characterized through a process called single-cell RNA sequencing.
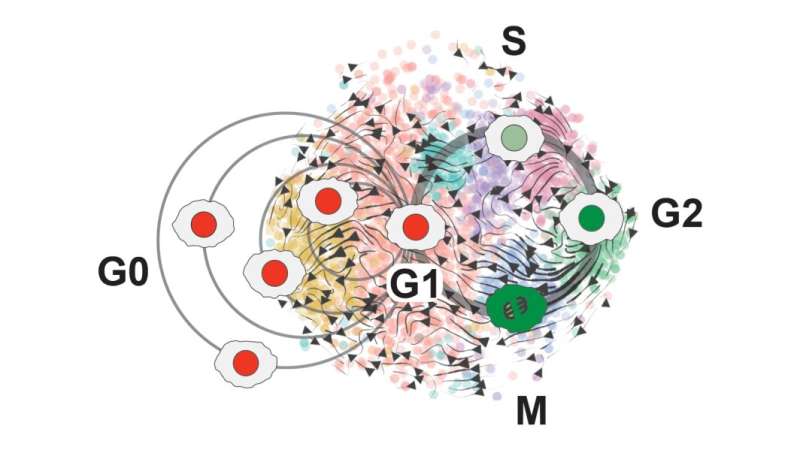
“That data turned out to be pretty amazing,” Plaisier says. “It mapped out into this beautiful circular pattern that we identified as all of the different phases of the cell cycle.”
O’Connor developed a new cell cycle classifier tool—called ccAF, or cell cycle ASU/Fred Hutchinson to represent the collaboration between the two institutions—that takes a closer, “high-resolution” look at what’s happening within the growth cycles of stem cells and identifies genes that can be used to track progress through the cell cycle.
“Our classifier gets deeper into the cell cycle because there could be pieces we’re capturing that have important implications for disease,” O’Connor says.
When Plaisier and O’Connor used the ccAF tool to analyze cell data for glioma tumors, they found the tumor cells were often either in the Neural G0 or G1 growth state. And as tumors become more aggressive, fewer and fewer cells remain in the resting Neural G0 state. This means more and more cells are proliferating and growing the tumor.
They correlated this data with the prognosis for patients with glioblastoma, a particularly aggressive type of brain tumor. Those with higher Neural G0 levels in tumor cells had less aggressive tumors.
They also found that the quiescent Neural G0 state is independent of a tumor’s proliferation rate, or how fast its cells divide and create new cells.
“That was an interesting finding from our results, that quiescence itself could be a different biological process,” Plaisier says. “It’s also a potential point where we could look for new drug treatments. If we could push more cells into that quiescent state, the tumors would become less aggressive.”
Current cancer drug treatments focus on killing cancer cells. However, when the cancer cells are killed, they release cell debris into the surrounding area of the tumor, which can cause the remaining cells to become more resistant to the drugs.
“So, instead of killing the cells, if we put them to sleep it could potentially be a much better situation,” Plaisier says.
With their ccAF tool, they were also able to find new states at the beginning and end of the cell cycle that exist between the commonly known states. These are among the topics for their next phase of research.
“We’re starting to think about ways to dig into those and learn more about the biology of the entry and exit from the cell cycle because those are potentially really important points where the cells will either go into the G1 state or G0,” Plaisier says.
Figuring out what triggers a cell to enter the division cycle or remain in a G0 resting state could help understand the processes behind tumor growth.
“The primary feature of any cancer is that the cells are proliferating,” Plaisier says. “If we could get in there and figure out what the mechanisms are, that might be a place to slow them down.”
Plaisier and O’Connor are making the ccAF classifier tool open source and available in a variety of formats for anyone studying single-cell RNA sequencing data to ease into the process of studying cell cycles.
Arizona State University

