In the face of cancer, oncologists face a dual epidemic of overtreatment and undertreatment. Treating the disease often involves harsh chemotherapies and other interventions with extensive side effects. As a result, clinicians might delay treating a growth that looks harmless until it becomes life threatening and too late to treat. Or, they might overly treat a less aggressive cancer with harsh therapies that cause more harm to the patient than benefit.
Now, two researchers, including Yale’s Jason Sheltzer, Ph.D., assistant professor of surgery (oncology), have conducted a study analyzing the molecular features of 33 different human cancers in an effort to find a way to differentiate between non-threatening and aggressive forms of the disease and found surprising results. They published their findings in Cell Reports on March 29.
“This is the first time that we have had a comprehensive understanding of the genetic differences between fatal and relatively harmless human cancers across several dozen cancer types,” says Sheltzer.
Biomarkers predict patients’ cancer outcomes
The identification of prognostic biomarkers in cancer may help clinicians assess which cancer patients are most at risk of developing aggressive forms of the disease. But while there is a lot of genomic data that has been collected for cancer research, translating that data into actionable clinical insights has been difficult.
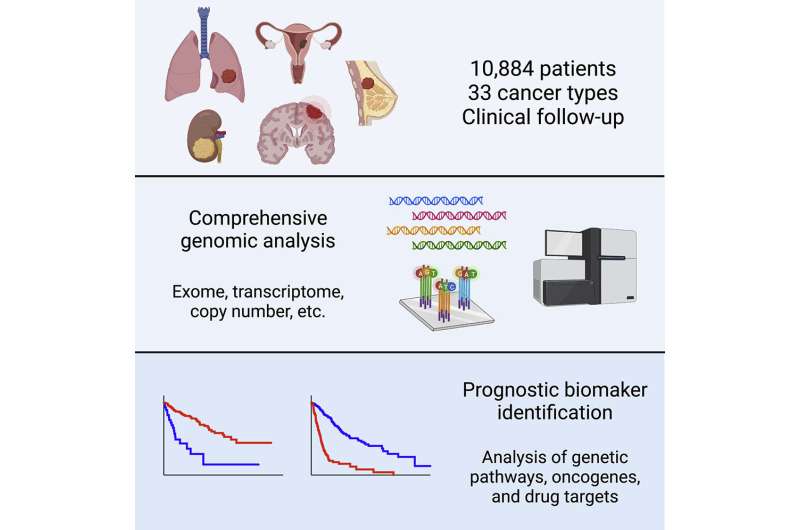
In 2006, the National Cancer Institute and National Human Genome Research Institute embarked on a large-scale, multi-year study called “The Cancer Genome Atlas Program.” The project recruited 10,884 patients with 33 different types of cancer and performed molecular profiling on the tumors from these patients. They analyzed data including the tumors’ gene expression, mutations, copy number alterations, DNA methylation, microRNA expression, and protein expression. Additionally, they collected information on the patients’ clinical course—whether they survived a few months or many years after surgery. “In our study, we linked these two pieces of data together,” says Sheltzer.
Alongside first author and wife Joan Smith, a senior staff software engineer at Google, Sheltzer assessed whether any of the genomic biomarkers present in cancerous tumors were related to shorter or longer survival times across patients. The study yielded a number of surprising results. For instance, cancer researchers have shown significant interest in using data on mutations from certain cancers as a potential prognostic biomarker, believing that patients whose cancers had certain mutations in a particular gene may experience worse outcomes and need to be monitored more closely. However, the team’s data suggest that mutations don’t have much prognostic significance in cancer.
Deaths associated with chromosome numbers
In contrast, they found that copy number alterations were highly prognostic and significantly associated with patient outcomes across multiple cancer types. Most cells in the human body have 23 pairs of chromosomes. In cancers, copy number alterations tend to occur when cells gain or lose chromosomes, leading to extra or fewer numbers of a gene. Sheltzer says his lab will pay particularly close attention to copy number changes in genes and chromosomes, and why they are associated with deaths from cancer, in its upcoming research.
“One surprising finding is that mutations conveyed relatively little information, but other types of genomic measurements from cancer, in particular copy number measurements, contained a lot of prognostic information,” says Sheltzer. Additionally, the team created a web portal that allows researchers to access the analyses they performed. They hope that their findings will help clinicians properly identify lesser cancers that don’t require intense treatments, as well as life-threatening cancers that require stringent intervention.
“Our work can potentially improve cancer patient well-being by saving the harshest treatments for those who need the most aggressive interventions,” says Sheltzer.
Isabella Backman, Yale University

