Using high-throughput genome sequencing and machine learning, scientists at UMass Chan Medical School have shown a strong correlation between the oral microbiome in patients with COVID-19 at the time of hospital admission and the need for later respiratory support. Published in Frontiers in Microbiology, the study adds to a growing body of research linking the oral microbiome with respiratory illnesses and offers new insights into how SARS-CoV-2 impacts inflammation and causes disease.
“Our results showed a correlation between the lack of certain organisms in the microbiome—specifically Prevotella salivae and Veillonella infantium—as well as a reduction in the abundance of metabolic pathways associated with LPS synthesis and the need for respiratory support after admission,” said principal investigator and co-author Evan S. Bradley, MD, Ph.D., assistant professor of emergency medicine and a member of the Program in Microbial Dynamics.
“This suggests that these microorganisms could play a protective role, perhaps by modulating pro-inflammatory pathways, in patients with COVID-19. We don’t know yet how this works on a biological level, but it offers a starting point for future investigations.”
Scientific evidence is emerging that the oral microbiome—the community of bacteria found in the upper tract—can influence the course of respiratory infections, Dr. Bradley said. Like the microbiomes found in the gut and on the skin, the oral microbiome is a collection of bacteria that can affect the progression of health and disease. Much smaller than the gut microbiome, the oral microbiome comprises a few hundred different species of bacteria. It is the first meeting place along the alimentary canal, the whole passage along which food passes from the mouth through the body, where the immune system meets the outside world.
Experts believe that maintaining the delicate balance of the oral microbiome can play an important part in health. An imbalance in the oral microbiome, like an imbalance in the gut microbiome, can lead to inflammation, illness and disease, according to the study authors.
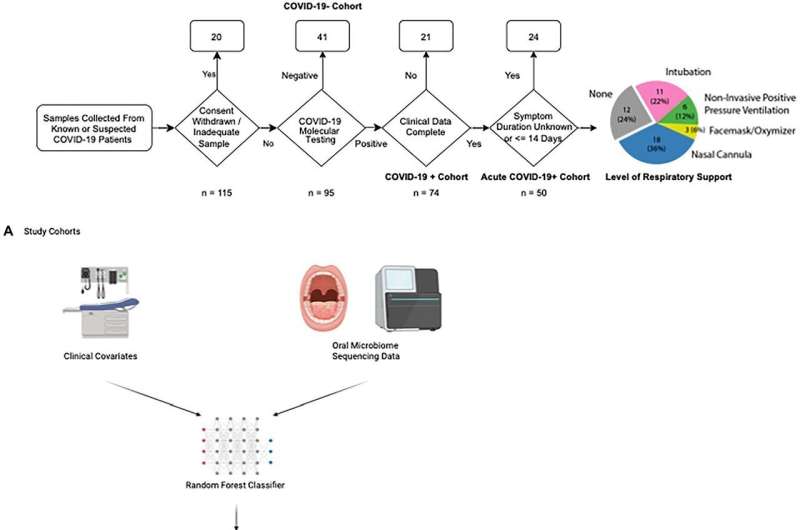
To understand the connection between the oral microbiome and COVID-19, Bradley sampled 115 patients presenting in the emergency room with COVID-19 symptoms. Blood and oral swabs were taken from patients, who were also tested for COVID-19 using PCR testing. Fifty of the 115 tested positive for COVID-19 and had symptoms for less than 14 days. Of this acute cohort, 38 required some form of respiratory support after admission.
Whole genome sequencing and advanced algorithms were used to identify the make-up of bacteria in the oral microbiome for each of the 115 samples in the cohort. To identify potential differences between the COVID-19 group that needed respiratory assistance and the one that did not, Vanni Bucci, Ph.D., associate professor of microbiology & physiological systems, employed machine learning based modeling to extract microbiome features, such as species and metabolites, that impact disease severity.
“Data from the microbiome is complex and multi-layered, so it’s not amendable to standard statistical analysis,” said Dr. Bucci, a member of the Program in Microbial Dynamics. “We have to use machine learning, which is like a black box. We input the features and data into the model. The model uses examples and controls to learn structures that it hasn’t seen before. From these structures, the model choses the best signal that fits the data. It tells you which features are extraneous, and which can’t be taken out.”
In Bucci’s model, low levels of the bacterial species P. salivae and V. infantium were predictive of the need for respiratory support in 85% of cases. Likewise, patients exhibiting a reduction in bacteria that produced metabolites associated with the LPS metabolic pathway had an 82% chance of needing respiratory support.
“We know inflammation is a major factor in COVID-19. It’s possible that these bacteria are somehow holding down runaway inflammation,” said co-author Abigail Zeamer, a Ph.D. student in the Department of Microbiology & Physiological Systems at UMass Chan.
The next step in the research, according to Bradley, is to inoculate an animal model with the bacterial species P. salivae and V. infantium and expose it to the SARS-CoV-2. “We can then start examining the relationship between these bacteria, inflammation and the immune system to understand how COVID-19 works.”
Jim Fessenden, University of Massachusetts Medical School

