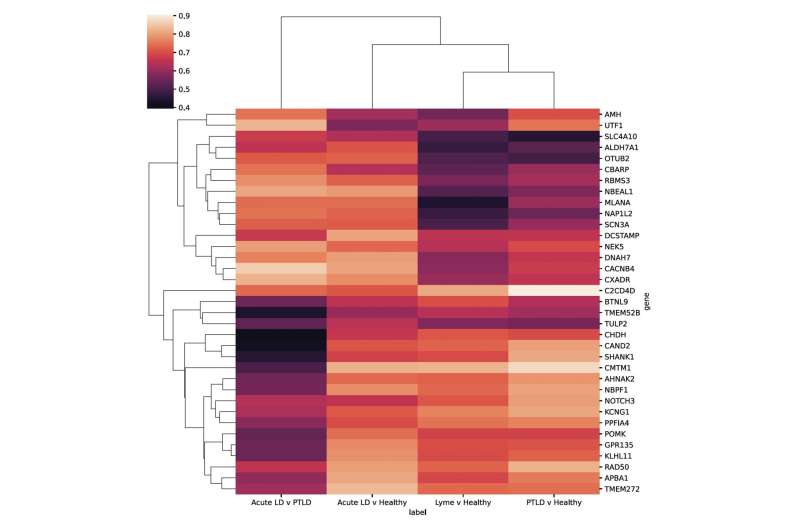
Researchers at the Icahn School of Medicine at Mount Sinai in New York have identified 35 genes that are particularly highly expressed in people with long-term Lyme disease. These genes could potentially be used as biomarkers to diagnose patients with the condition, which is otherwise difficult to diagnose and treat.
The findings, published November 15 in the journal Cell Reports Medicine, may also lead to new therapeutic targets. The study is the first to use transcriptomics as a blood test to measure RNA levels in patients with long-term Lyme disease.
Lyme disease is a tick-borne illness that is not well understood. Approximately 30,000 diagnosed cases are reported to the CDC each year, but the estimated real number is closer to 476,000 cases, carrying an annual healthcare cost of about $1 billion in the United States. While most patients are diagnosed and treated with antibiotics at the earliest stages of Lyme disease, about 20 percent of the patients develop long-term complications, which could include arthritis, neurologic symptoms, and/or heart problems.
“We wanted to understand whether there is a specific immune response that can be detected in the blood of patients with long-term Lyme disease to develop better diagnostics for this debilitating disease. There still remains a critical unmet need, as this disease so often goes undiagnosed or misdiagnosed,” said Avi Ma’ayan, Ph.D., Professor, Pharmacological Sciences, and Director of the Mount Sinai Center for Bioinformatics at Icahn Mount Sinai, and senior author of the paper. “Not enough is understood about the molecular mechanisms of long-term Lyme disease.”
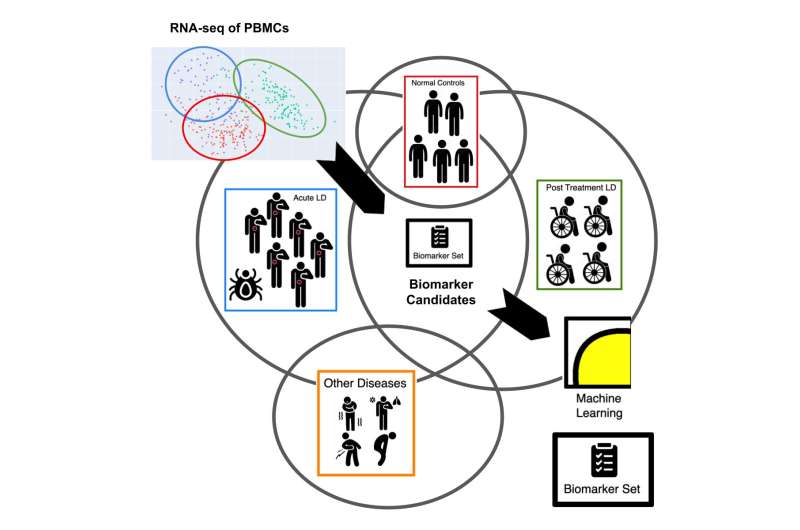
As part of the study, RNA sequencing was conducted using blood samples from 152 patients with symptoms of post-treatment Lyme disease to measure their immune response. Combined with RNA sequencing data from 72 patients with acute Lyme disease and 44 uninfected controls, the investigators observed differences in gene expression and found that most of the post-treatment Lyme disease patients had a distinctive inflammatory signature compared with the acute Lyme disease group.
In addition, by analyzing the differentially expressed genes in this study along with genes that are differentially expressed due to other infections from other published studies, the researchers identified a subset of genes that were highly expressed, which have not been previously established for this Lyme-associated inflammatory response.
Using a type of artificial intelligence called machine learning, the researchers further reduced the group of genes to establish an mRNA biomarker set capable of distinguishing healthy patients from those with acute or post-treatment Lyme disease. A gene panel that measures the expression of the genes the investigators identified could be developed as a diagnostic to test for Lyme.
“We should not underestimate the value of using omics technologies, including transcriptomics, to measure RNA levels to detect the presence of many complex diseases, like Lyme disease. A diagnostic for Lyme disease may not be a panacea but could represent meaningful progress toward a more reliable diagnosis and, as a result, potentially better management of this disease,” said Dr. Ma’ayan.
Next, the investigators plan to repeat the study using data from single-cell transcriptomics and whole blood, apply the machine learning approach to other complex diseases that are difficult to diagnose, and develop the diagnostic gene panel and test it on samples from patients.
The paper is titled “Gene set predictor for post-treatment Lyme disease.” Additional co-authors are Daniel J.B. Clarke, MS (Icahn Mount Sinai, New York), and Alison W. Rebman, MPH, Jinshui Fan, MD, Ph.D., Mark J. Soloski, Ph.D., and John N. Aucott, MD, all from Johns Hopkins University of Medicine in Baltimore.
The Mount Sinai Hospital

