Researchers in the Muir Lab at Princeton University’s Department of Chemistry have completed the first comprehensive analysis of cancer-associated histone mutations in the human genome, featuring both biochemical and cellular characterizations of these substrates. Their study reports that histone mutations that perturb nucleosome remodeling may contribute to the development or progression of a wide range of human cancers.
Within the human genome, DNA is wrapped around disc-shaped structures made up of eight histone proteins, each forming nucleosomes. Repeating nucleosome units comprise chromatin, a storehouse of genetic information that is both structured and dynamic. Broadly, the Muir Lab seeks to understand how chromatin controls genetic processes in the cell and how disruption of these processes may cause disease.
The new research, “Oncohistone mutations enhance chromatin remodeling and alter cell fates,” was reported this week in Nature Chemical Biology. Members of the Allis Lab at The Rockefeller University, long time partners of the Muir Lab, were key collaborators in the work.
The research builds on a paper published by the Muir and Allis labs last year in Nature.
“We noticed based on that previous work that a lot of different mutations in histones were associated with different cancers and to different degrees,” said Michelle Mitchener, one of the paper’s lead authors and a postdoc in the Muir Lab. “That previous paper was kind of an overview of where the mutations are located in chromatin and hypotheses about what they might be doing. But it was more of a data-mining effort.
“So, this paper was focused on trying to figure out functionally and biochemically what those mutations are actually doing,” she added. “If they contribute to cancer, then how? Can we figure out at a structural and biochemical level what they’re doing?”
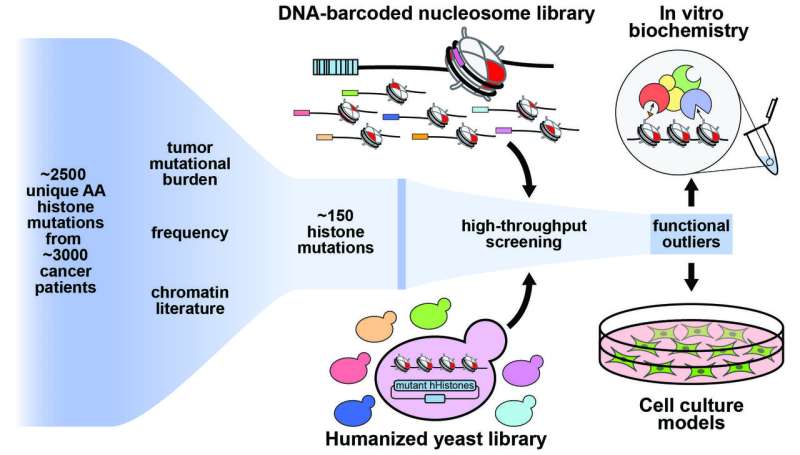
As often happens in the practice of science, the researchers generated new tools for investigating these “oncohistones.” They created two experimental libraries containing highly relevant histone mutations to profile their biological effects. The libraries—one biochemical and one in yeast—will enable researchers in future projects to investigate these biological processes more quickly and efficiently.
In this particular investigation, researchers looked at mutations within the cores of the histones themselves to see if and how they might be impacting disease states.

“We found mutations inside the core of the histones—in all four histones—and they seem to do completely different things than previously identified histone mutations,” said John Bagert, the co-lead author on the paper, also a postdoc in the Muir Lab. “They affect the actual structure of the nucleosome itself and seem to destabilize it in some cases.
“These are fundamental processes that people studying chromatin think about all the time, but they have not been linked in this way to cancer previously,” Bagert added. “So, we think that mutations that affect chromatin remodeling can contribute to disease and cancers in humans. We’ve identified the sites and the mutations at those sites that we think are causing problems.”
Multiple mutations converged
The three broad sites in the nucleosome structure where mutations had the most pronounced effects include: the DNA histone interactions; a specific binding site called the acidic patch; and a key structural interface within the folded nucleosome structure that affects its stability. Multiple mutations converged on these sites, raising the possibility that they are hotspots for disease development.
Researchers emphasized that some of these mutations are showing up very early in the disease cycle. Whether they turn out to be contributive is among the most pressing questions in cancer research.
“We do see that some of these mutations show up very early,” said Mitchener. “At what stage? We just don’t know yet. But part of me is most excited that we started the process of figuring out what these mutations do. We identified some new sites that I think the field should go after and study more. There are still lots of mechanistic questions.”
For his own part, Bagert said he looks forward to the ways their libraries and other high throughput methods can enable biochemical investigations in the future.
“For decades now, we have had really robust methods of doing biochemistry and chemistry in biological systems, but it’s kind of been a one-Ph.D.-one-protein type of approach. But now, many labs along with ours are trying to come up with ways that we can answer all the questions at once, just by being smart about our approach.
“Speed is everything. There’s so much biology out there and we cannot do it all,” he said. “So we just need to come up with ways of doing it fast. I think that this is the future of biology.”
Princeton University

