COVID-19, in its multiple variants and its ability to thwart efforts to wipe it out, still has a lot of unknowns that make it impossible for scientists to declare victory over the disease, despite vaccines.
But now two Université de Montréal professors working at the Montreal Institute for Clinical Research (IRCM) have taken a big step towards understanding the coronavirus: they’ve identified and confirmed the power of two small molecules to block the infection of lung cells by SARS-CoV-2, the virus that causes COVID-19.
The finding by the teams of medical professor Nabil G. Seidah, the IRCM’s director of biochemical neuroendocrinology research, and microbiology professor Éric A. Cohen, the IRCM’s director of human retrovirology research, was published Monday in the Journal of Virology.
How it works and why
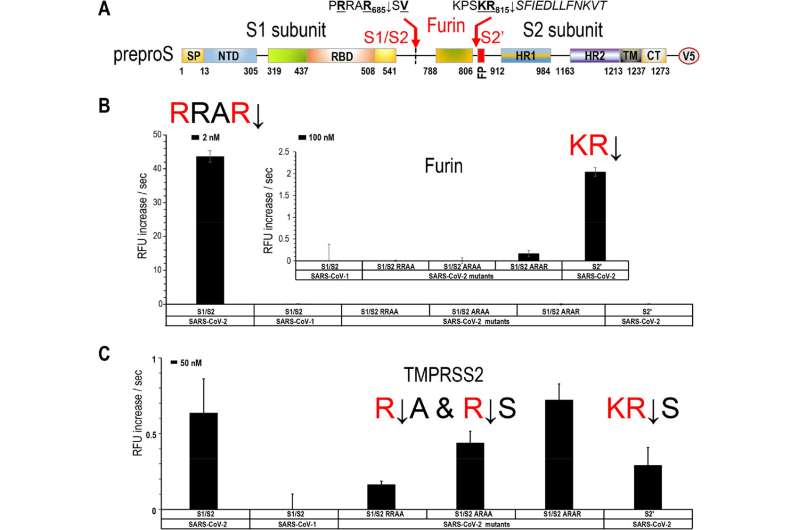
Viral entry into lung epithelial cells requires cleavage (↓) of the viral surface spike-glycoprotein of SARS-CoV-2 at two sites, S1/S2 (PRRAR↓) and S2′ (KPSKR↓) to expose a fusiogenic sequence allowing host membrane fusion with the infectious virus.
Seidah’s team was already the first to predict that the 4-residue insertion (PRRA) in the spike sequence would result in a typical proprotein convertase furin-like cleavage at the S1/S2 sequence of PRRAR↓.
The new study validates this concept and shows unequivocally that furin cleaves at both sites, enhancing viral infection, the scientists say. They also show that another enzyme, TMPRSS2, cleaves the membrane-bound receptor ACE2 and releases it into the medium, promoting cellular entry of SARS-CoV-2.
The result: combining of potent small molecule inhibitors of furin (BOS compounds) and TMPRSS2 (Camostat) blocks the live viral infection of lung cells by more than 95 percent.
Another weapon to fight COVID-19
SARS-CoV-2, the etiological agent of COVID-19, has caused more than 6 million deaths worldwide, and that number is still rising. Seidah and Cohen say their powerful new antiviral approach will help reduce the spread of SARS-CoV-2.
That’s especially important, they add, as other coronaviruses are widely expected to develop in the near future. Having new treatments in the antiviral arsenal, they say, will help society better prepare for future pandemics.
University of Montreal

